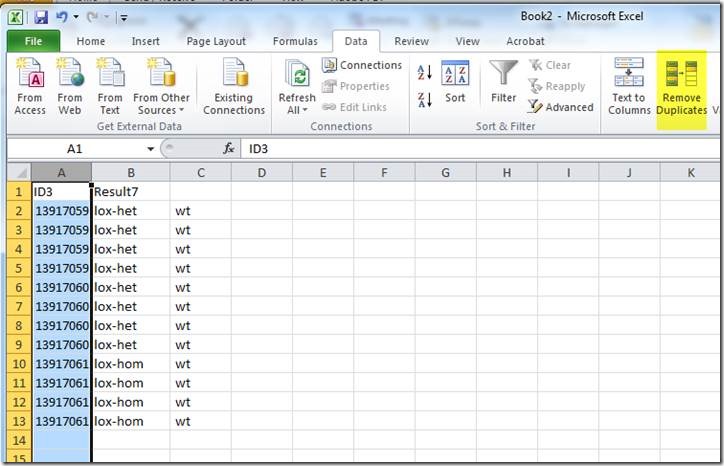
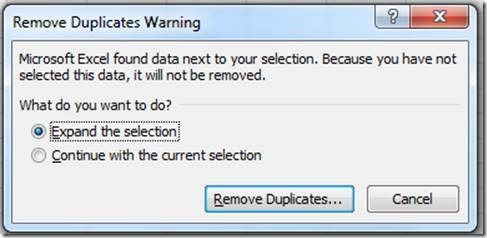
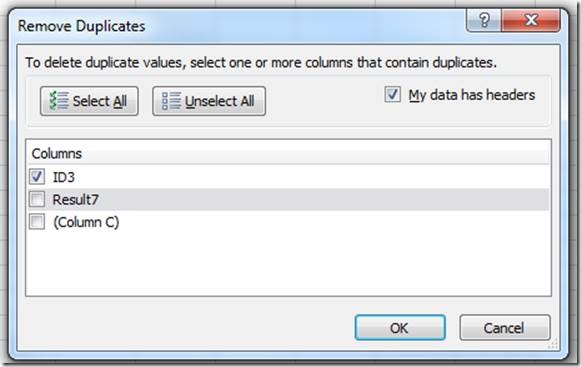
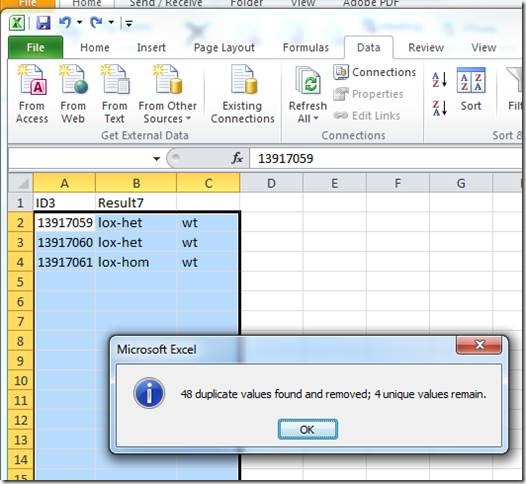
A mating record in Mosaic Vivarium represents a list of possible parents for a set of litters. When you put animals together, Vivarium will inspect the list of existing mating records for the same set of parents – if the exact same set of parents exists for a mating record, Vivarium will re-use that mating record for the new mating cage. That means new litters seen in the cage will be added to the existing mating record.
If the set of possible parents in a cage has not been seen together before, a new mating record is created. New litters seen in the cage will be added to the new mating record.
The mating record serves at the basis of your pedigree. To find all ancestors of an animal, Vivarium inspects the parents associated with the mating ID from which the animal came. Then Vivarium looks at the mating IDs from which those parents came, etc., until it finds animals from the “N/A” mating ID. Those animals are ones which were received into the system (not bred in the facility).
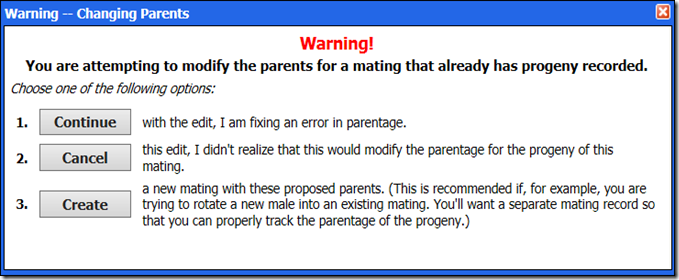
IMPORTANT: The only reason to modify the parent animals associated with a mating ID is to correct an error in the identity of one or more of the parents. When you attempt to make such a change, you will see the following message:
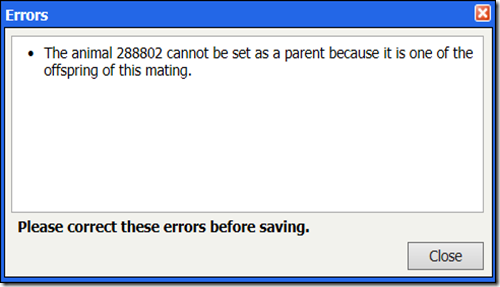
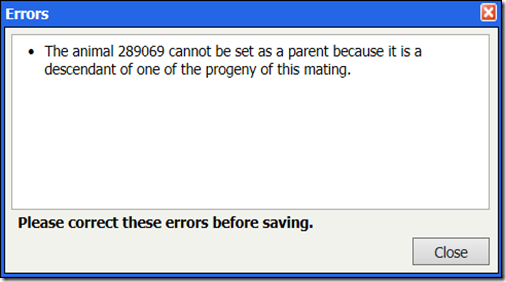
In rare cases, we have seen cases of people modifying a mating record and recording a descendant as a parent. If you do this, you will see one of the following two messages:
or:
Some earlier versions of Mosaic Vivarium did not perform this check for you, and it was possible to make an error leading to an impossible pedigree (like me being my own grandfather). We have seen one or two instances of this out of hundreds of thousands of matings.
If you do have a pedigree loop, you will see an error when you try to setup a mating indicating the loop. The specific animal will be identified. The easiest way to fix this is:
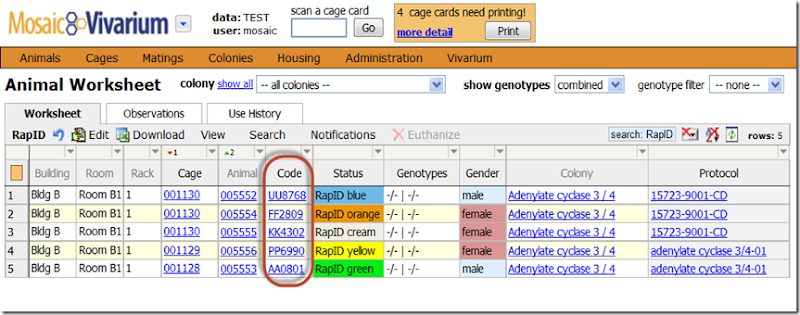
- Search for the animal on the Animal Worksheet.
- Right-click on the animal’s row, and choose Mating –> “All Matings with these Parents”.
- Next, remove that animal from the list of parents in oldest mating record.
All your animal records will remain, but you will have incomplete pedigree information for some animals. Of course the pedigree is already incomplete since this error likely occurred by replacing a correct parent in the mating record with a new one.
The important thing to understand is that setting up new matings is the process of putting a different set of possible parents together, resulting in a new mating ID. Existing mating records which have litters recorded are the birth records for those animals. Changing the parents in the offspring’s birth records (mating ID) only makes sense as an error correction if the original parents were misidentified.