The following information is provided courtesy of Dr. Christian Schmedt of the Genomics Institute of the Novartis Research Foundation.
Tables with genotype data linked to specific mouse IDs can be uploaded to Mosaic via the "Genotyping" menu under "Animals":
Uploading genotypes requires a comma-separated-value (csv) file listing the animal ID in column A and genotypes in the adjacent columns (B, C, maybe D). The genotypes need to be expressed the same way as the long genotype name defined under Administration>Genotypes.
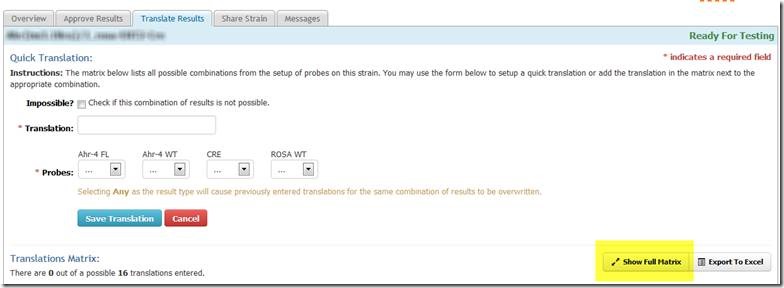
In order to derive a genotype upload file from Transnetyx results, the genotyping assay on the Transnetyx site first needs to be translated to reflect long genotype names in Mosaic:
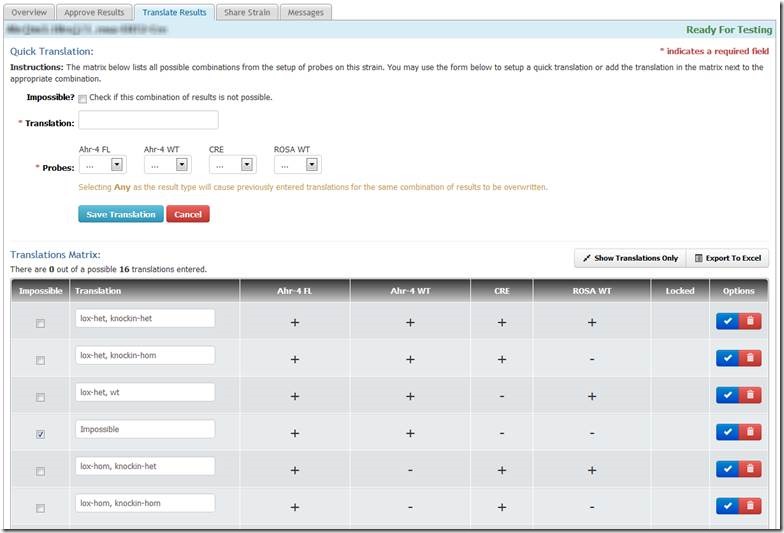
Translation is easiest in the "full matrix" mode:
(matrix shown is incomplete)
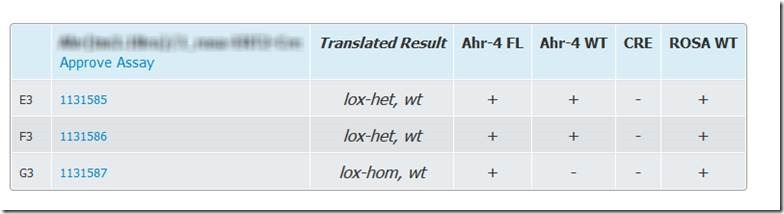
Translation will be reflected in the way the results are displayed on the Transnetyx website:
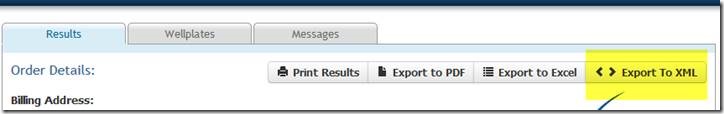
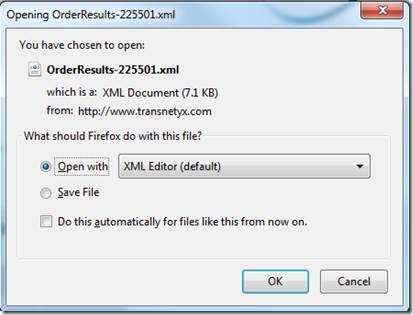
In order to extract this information and process for Mosaic genotype upload, export the data as XML file:
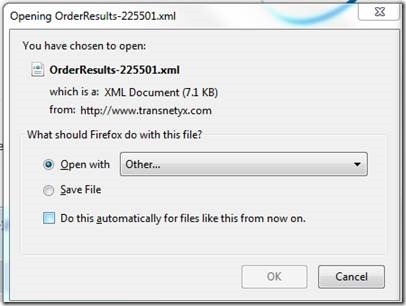
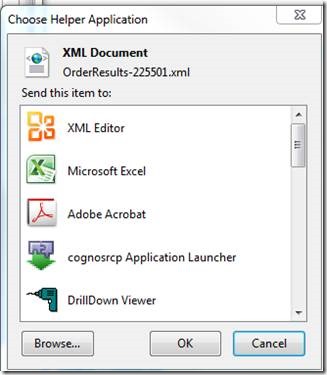
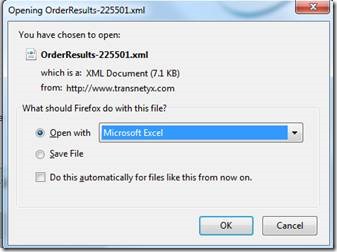
Switch from "XML editor" to "Excel" to open the document:
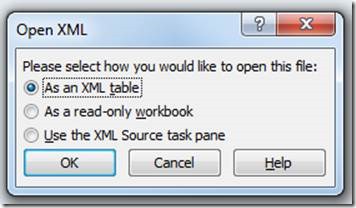
And open as XML table in excel:
Click "OK" on the next two dialogs:
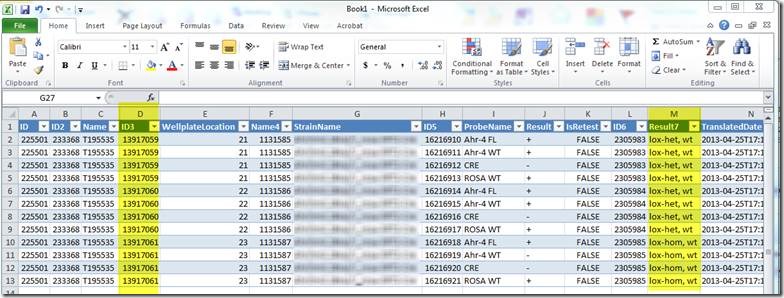
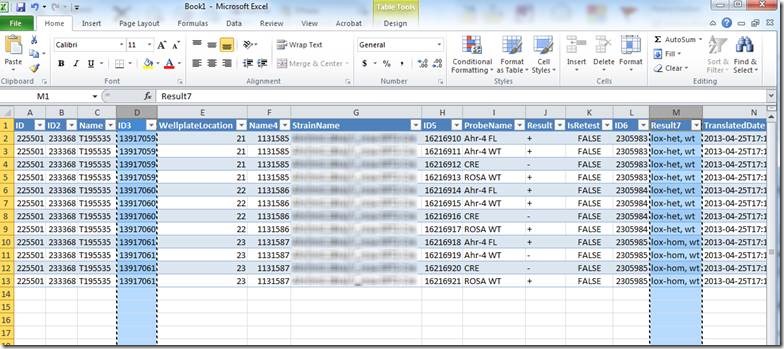
The resulting Excel document looks like this:
Column "D" contains the animal ID (this may be different for different Transnetyx users, but the animal ID will be in one of the columns). Column "M" contains the translated result.
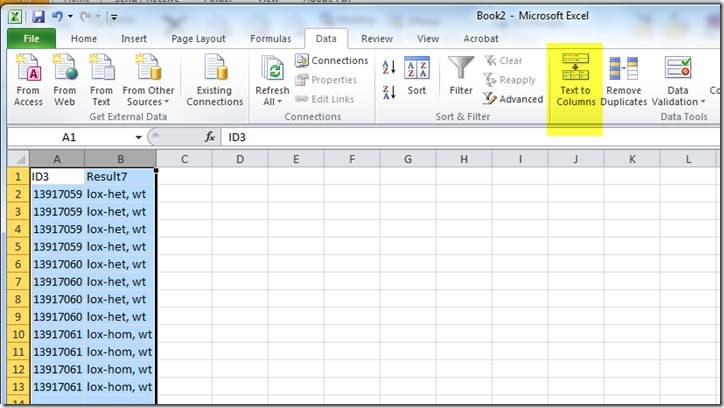
Copy columns D and M to a new excel document.
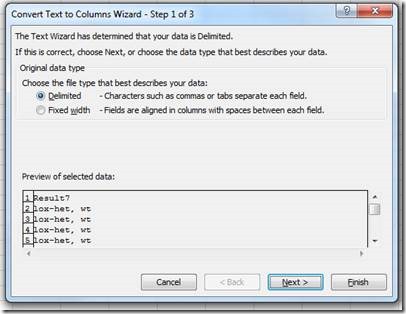
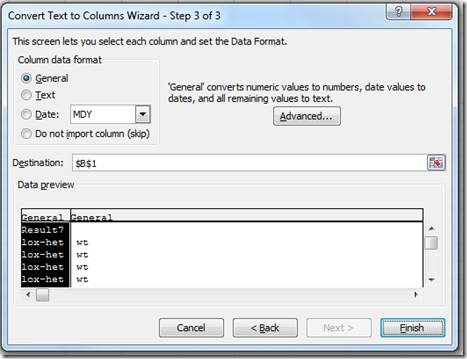
In the new document, use the "Text to Columns" tool to separate the translated results into two columns:
Choose delimited
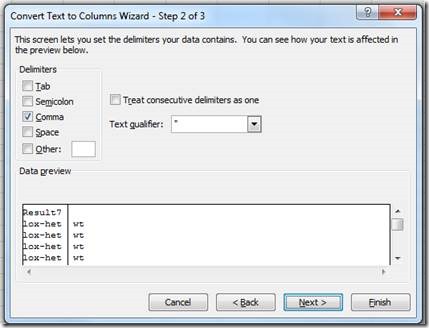
"Comma" (this may be Space or something else, dependent on how the translations were defined for the Transnetyx assay)
And finish
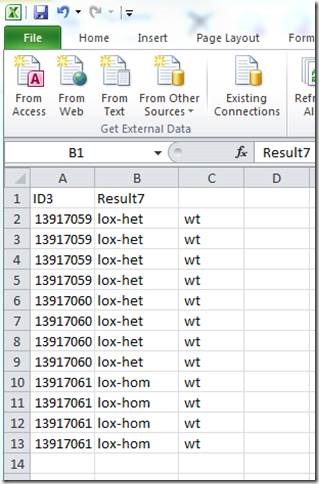
To get two columns for genotypes:
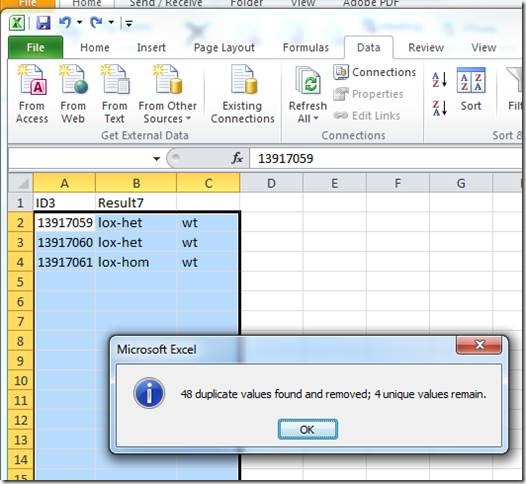
Note that each ID is represented by 4 rows (one for each Transnetyx probe-set).
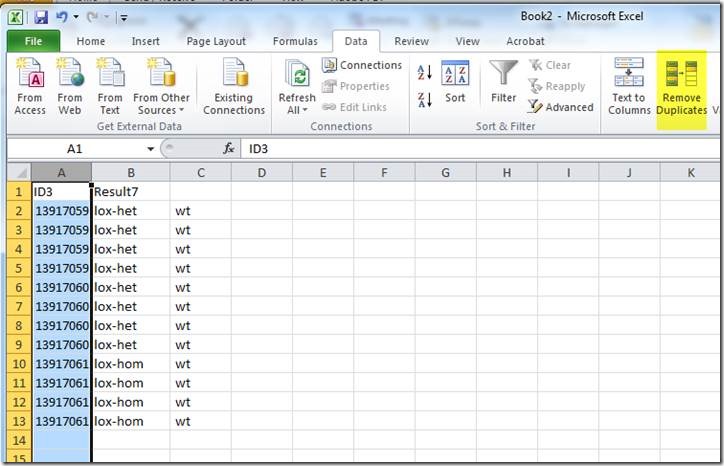
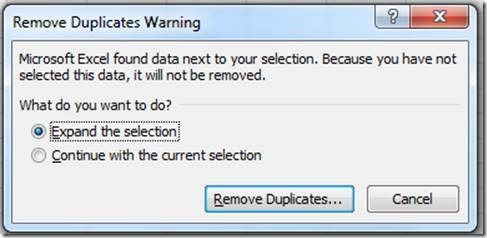
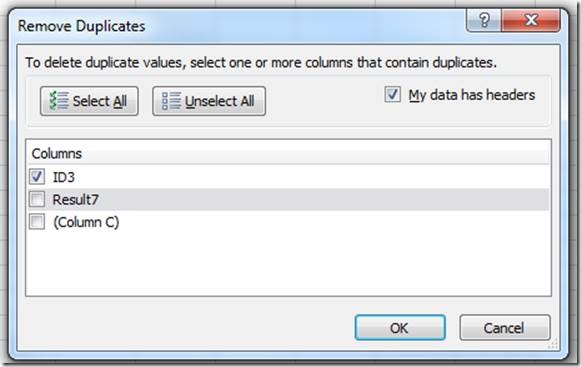
Before saving the file, the duplicates need to be removed with the "Remove Duplicates" tool:
Expand the selection beyond column A, which has the animal IDs
Unselect the columns with genotypes:
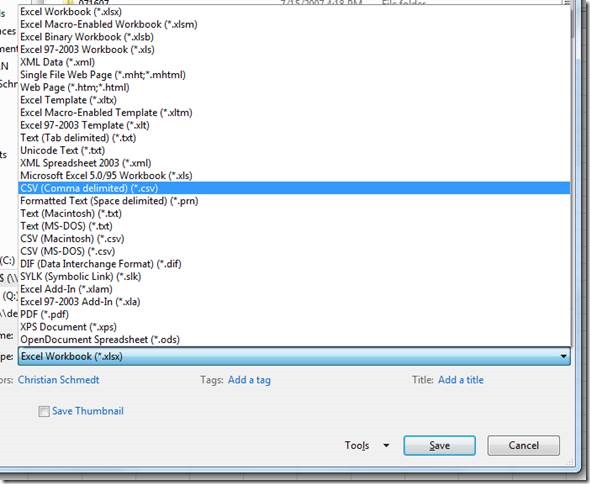
Click "ok" and save the files as csv:
The saved file is ready for upload into Mosaic.